 Research My scientific research interests are mainly in the field of biomolecular NMR. I study the structure and dynamics of proteins, RNA, antibiotics and their complexes using NMR spectroscopy. Therefore I work on several projects in which the biological targets range from small organic molecules to large biomolecular complexes. These studies are very diverse and contain many aspects of the modern biomolecular NMR; ranging from protein expression in E-coli, in-vitro synthesis of RNA, recording, analysing and assigning NMR spectra to calculation of the structure. Below are some examples of my research projects.  Study of the Cdc37-Hsp90 complex The molecular chaperone proteins Cdc37 and Hsp90 are crucial elements in the protein signalling pathway. The catalytic domains of many kinases are stabilized by Cdc37 and their proper folding and functioning is dependent on Hsp90. The X-ray crystal structure of the 16 kDa middle domain of human Cdc37 was solved at 1.88 Å resolution. Using NMR data and docking we determined the structure of this domain in complex with the 23 kDa N-terminal domain of human Hsp90. The results demonstrate that the middle domain of Cdc37 exists as a monomer and reveal the key residues that enable complex formation. These findings can be very useful in the development of small molecule inhibitors against cancer.  NMR structure of the alpha-amylase inhibitor Parvulustat The protein parvulustat (78 amino acids) functions as a highly efficient alpha-amylase inhibitor and has 29.6% sequence identity to the well-known alpha-amylase inhibitor tendamistat. The high-resolution NMR structure of parvulustat was solved and its dynamical properties were studied. The overall structural composition is very similar to tendamistat, but there are distinct differences including the active-site region which indicates a slightly different binding mode. We propose that an induced-fit mechanism may take place for binding to alpha-amylase since the structure of tendamistat does not change upon binding to alpha-amylase.  Deployment and integration of a NMR e-infrastructure Biomolecular NMR plays an important role in life sciences, and structural biology in particular. An EU-NMR initiative is already operative in this field, providing access to modern NMR instrumentation and pursues technical advancement. Furthermore, a Coordination action NMR-Life aims at the establishment of the best common experimental methods and to spread them across Europe. In addition the new initiative, eNMR, endeavors deploying and unifying the NMR computational infrastructure. The project is funded under the 7th framework programme of the European Union and its main objective is the implementation of a platform for computational approaches necessary for biomolecular NMR data analysis. This eNMR network will be based on the Grid (EGEE) infrastructure. 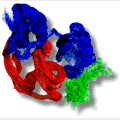 L11 domain rearrangement upon binding to RNA and thiostrepton Conformational dynamics have been studied for the ribosomal protein L11 (15.1 kDa, 141 aa) in its free form as well as in the highly conserved complex with the GTPase-associated region of the 23S rRNA (19.3 kDa, 60 nt) and in the ternary complex with the thiazole antibiotic thiostrepton. The RNA is compactly folded into a well defined tertiary structure, which is further stabilized by association with the C-terminal domain of L11 (L11ctd). NMR data revealed a rearrangement of the N-terminal domain of L11 (L11ntd), placing it closer to the RNA after binding of thiostrepton, which may prevent binding of elongation factors and consequently disrupts the elongation factor function. A model is proposed for the ternary L11-RNA-thiostrepton complex that is additionally based on interaction data and conformational information of the L11 protein providing an explanation for the role of L11ntd in elongation factor binding. 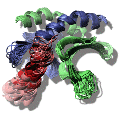 Interactions between the cofactor PC4 and activator VP16 These studies demonstrated how the intrinsically unstructured domains of PC4 and VP16 are involved in the regulation of RNA Polymerase II transcription. The highly flexible N-terminal domain of PC4 (PC4ntd) is mostly involved in the transient and dynamic regulation of the PC4 cofactor function and is controlled by phosphorylation. The activation domain of VP16 (VP16ad) adopts an alpha helical structure upon binding to PC4 and TFIIB and the docking models provide an explanation for the high activity, cooperativity and promiscuous properties of this transactivation domain. The structural and functional studies of PC4, VP16 and the phosphorylation of PC4 lead to a putative model that explains the different cofactor activities of PC4 in the context of the transcription cycle.  |