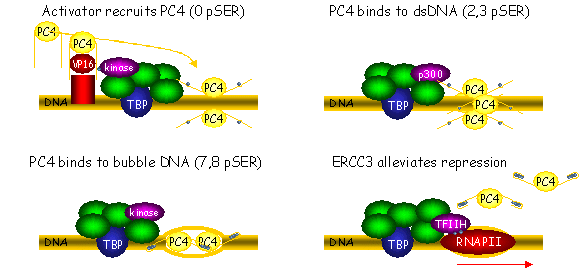
Summary Thesis Henry Jonker: PC4 and VP16 Understanding the molecular level of gene expression gives us insight in the connection between genotype and phenotype. This requires investigation of both bio-functional and structural aspects of many factors involved in this process. Of specific interest is the precise and specific regulation of RNA Polymerase II transcription. Transcriptional activators and cofactors can bind to particular DNA regions and basal transcription factors in order to enhance or repress transcription. This thesis deals with structural biology of the general transcriptional cofactor PC4 and the activator VP16. Previous work on the PC4 protein revealed a bipartite structure. The carboxy terminal domain of PC4 (PC4ctd) is involved in high affinity binding to single-stranded DNA (ssDNA), preferentially to two strands running in opposite direction as for a DNA bubble. PC4 stabilizes the pre-initiation complex (PIC), targets many transcription factors and has the ability to unwind DNA. The dimeric structure of PC4ctd had been solved earlier by crystallography and studied by NMR. However, little was known about the unstructured amino terminal domain of PC4 (PC4ntd), which is involved in protein-protein interactions and binding to dsDNA. Furthermore, activation domains are frequently unstructured and it was mostly unknown how they interacted with the other proteins of the transcription machinery. The aim of this study is to obtain structural and functional information about the interaction of PC4 with the activation domain of VP16 (VP16ad), using high-resolution NMR spectroscopy and biochemical analysis. Furthermore, the phosphorylation of PC4 is investigated, which has previously been reported to influence the cofactor function. Initial NMR analysis of PC4 (Chapter 2) revealed that the VP16ad interaction site of PC4 partially overlaps with the ssDNA binding site on PC4ctd. Site directed mutagenesis largely confirmed the binding site and revealed a quantitative correlation between PC4ntd residues that stimulate the interaction with VP16ad and negatively affect the ssDNA binding and DNA unwinding. Biochemical experiments demonstrate that VP16ad blocks the DNA unwinding activity and indicate competitive binding of VP16ad and DNA to PC4. Most interestingly, PC4ctd is affected by the presence of PC4ntd, suggesting a regulatory role for PC4ntd. This domain may modulate and/or shield the functional surface in PC4ctd to adjust the binding affinity to either activators or ssDNA. PC4ntd comprises a region rich in serine and acidic residues (SEAC), a lysine rich region and another serine rich region. Structural studies showed that PC4ntd is highly flexible and mostly unstructured, although the lysine rich region, and flanking residues, exhibits a tendency to form secondary structure elements. These characteristics did not significantly change upon interaction with VP16ad, indicating that a rigid tertiary structure of PC4ntd is not formed. However, by NMR, chemical shift changes were observed in the lysine rich region and the SEAC region of PC4ntd, indicating that it participates in the interaction. As the interaction is mostly electrostatically driven, the lysine rich region of PC4ntd can increase the on-rate of VP16ad binding by providing an additional positively charged surface. Furthermore, the flexibility is somewhat reduced in the first a-helical part of the lysine rich region, indicating that it may also decrease the VP16ad off-rate by stabilizing the PC4ctd-VP16ad interaction. The NMR and biochemical data indicate a dynamic regulatory role for PC4ntd to regulate the PC4 activities by transient interactions with PC4ctd, activators and dsDNA. The SEAC region of PC4ntd can be phosphorylated, which has important functional consequences. The mode of phosphorylation, and the crucial impact on the cofactor function of PC4 have been studied in detail using biochemical analysis, MS and NMR (Chapter 3). The biochemical properties of PC4 are shown to be influenced in a distinct phosphorylation status dependent fashion. One phosphoserine is already sufficient to increases ssDNA binding affinity, which does not change upon further phosphorylation. The presence of more than one phosphoserine causes a loss of VP16 binding and a decrease of the DNA unwinding activity. The dsDNA binding affinity is gradually decreased to a minimum when phosphorylation is complete. Significant chemical shift changes are observed in the b-sheet regions of PC4ctd upon phosphorylation, that are most probably the result of a changed environment due to temporary interactions between PC4ctd and PC4ntd. Analysis of MS and NMR data revealed that up to eight serines are progressively phosphorylated towards the amino-terminus of the SEAC region, resulting in gradual chemical shift changes in the carboxy-terminal direction of the first part of the lysine rich region. As the affected region is involved in the interaction with activators and DNA, it is supposed that the functional lysine rich region is gradually masked by the phosphorylated SEAC region leading to coordinated changes in the PC4 cofactor function. The regulated phosphorylation has been correlated with the cofactor activities of PC4 that are performed during the transcription cycle. Finally, the properties of VP16ad are studied (Chapter 4). NMR experiments indicate that two regions are involved in the interaction with PC4 and undergo a conformational transition from random coil to a-helix upon binding. The same regions are affected in a similar way upon binding to the carboxy terminal domain of the general transcription factor TFIIB (TFIIBc). Biochemical experiments revealed that both activation subdomains are required for cooperative interaction with PC4 and confirm that the interaction is strongly electrostatically driven. Models for the PC4ctd-VP16ad and TFIIBc-VP16ad complexes are presented, based on NMR chemical shift changes and two independent docking approaches. The models are consistent with results from site directed mutagenesis. This study demonstrates how the intrinsically unstructured domains of PC4 and VP16 are involved in the regulation of RNA Polymerase II transcription. The highly flexible PC4ntd is mostly involved in the transient and dynamic regulation of the PC4 cofactor function, which is controlled by phosphorylation. VP16ad adopts structure upon binding to various targets and the docking models provide an explanation for the high activity, cooperativity and promiscuous properties of this transactivation domain. The structural and functional studies of PC4, VP16 and the phosphorylation of PC4 lead to a putative model that explains the different cofactor activities of PC4 in the context of the transcription cycle (Fig. 1). PC4 is first recruited to the promoter region through interaction with activators (VP16 Kd ~ 1 ÁM, Chapter 4). Phosphorylation of a few serines by kinases, present in the PIC, decreases the affinity for the activator. At this intermediate phosphorylation state (2-3 phosphoserines), PC4 binds to dsDNA (Kd ~100 nM, Chapter 3). Further phosphorylation will reduce the dsDNA binding affinity. PC4 can, most probably in cooperation with other transcription factors, unwind dsDNA and bind to the DNA bubble (Kd ~10 nM, Chapter 3) thereby stalling transcription. When the PIC assembly is complete, the presence of the ERCC3 component of TFIIH alleviates PC4 repression to enable initiation of transcription. PC4 can inhibit the phosphorylation of RNA Polymerase II (RNAPII), an event generally thought to be a critical step in the conversion from transcription initiation to elongation. As phosphorylated PC4 is no longer able to inhibit the phosphorylation of RNAPII, the transcription process may proceed to the elongation phase of transcription.
 |