 |
||||||
| Articles | Projects | Resume | Cartoons | Windsurfing | Paintings | Album |
Microbiological Analysis of the Nitrogen Cycle:
Isolation and Identification ofRhizobium andAzotobacter
Nitrate Reduction
by Waterose
Index:
Figures:
- Gram Stain Drawing of Rhizobium Bacteria Cells from Crushed Nodule on Day 1.
- Gram Stain Drawing of Rhizobium Bacteria Cells from colony on Rhizobium Agar Plate incubated at 30 Degrees Celcius for 3 Days.
- Gram Stain Drawing of Azotobacter Bacteria Cells from colony on Mannitol N-free Agar plate incubated at 30 Degrees Celcius for 3 Days.
Tables:
Table 1. Cell Morphology of Rhizobium Cells from Freshly Crushed Nodule on Day 1.
Table 2. Colony Morphology of Rhizobium .
Table 3. Cell Morphology of Rhizobium Cells from colony on Rhizobium Agar plate incubated at 30° C for 3 days.
Table 4. Colony Morphology of Azotobacter obtained from the Isolation in Flask.
Table 5. Cell Morphology of Azotobacter Cells from colony on Mannitol N-free Agar plate incubated at 30° C for 4 days.
Table 6. Nitrate Reduction Tests on Bacteria in Nitrate Broth in Durham Tubes incubated at 37° C for 7 days.
The nitrogen cycle is an biogeochemical essential process that converts different forms of nitrogen and compounds of nitrogen into forms that can be utilized for life processes. The limiting factor is that atmospheric nitrogen (N2) is unavailable to most organisms due to the high amount of energy required to break the triple bond between the two atoms of nitrogen. Alternatively, other forms of nitrogen, such as ammonia (NH3+), is converted to soluble forms of nitrogen compounds, including nitrites and nitrates that can be utilized. The utilisation of nitrogen is a complex process that involves many different chemical reactions by different organisms.
Nitrogen fixation is the process of converting nitrogen gas to a fixed form which is a nitrogenous compound, such as ammonia (NH3) or ammonium (NH4+). The reaction generates hydrogen gas, which can be utilized to reduce molecular oxygen and generate electrons and ATP. The equation for this reaction in most nitrogen fixing organisms, such as Rhizobium and Azotobacter is :
Equation 1. Nitrogen Fixation.
![]()
The net reaction after the H2 is recycled is:
Equation 2. Net Nitrogen Fixation Reaction.
![]()
The ammonia/ammonium is converted by nitrification to nitrite and nitrate, which are utilized by the bacteria and the plants for organic life processes. The equations for this two step reaction are:
Equation 3. Nitrification: Oxidation of ammonium to nitrite.
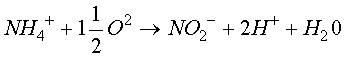
Equation 4. Nitrification: Oxidation of nitrite to nitrate.
![]()
The soluble form of nitrite and nitrate can be assimilated by plant roots and utilized in synthesising proteins and acids. This form of nitrogen can be converted to ammonia by plants, animals and micro-organisms. Animals return nitrogenous wastes to the environment as uric acids (Atlas, 1998).
The reverse process of nitrification is denitrification which reduces nitrates to nitrogen gas, returning the N2 gas to the atmosphere and the starting point in the cycle as described here. The denitrification can be accomplished by specific organisms, including the Pseudomonas, the Azospirillum, and the Rhizobium (Atlas, 1998). The equation that describes this multi-step reaction is:
Equation 5. Denitrification Multi-step reaction.
![]()
The process of denitrification uses the nitrate as an alternative to atmospheric oxygen as an electron acceptor for anaerobic respiration.
Materials and Methods are as outlined in the Experiment 12: Aspects in the Nitrogen Cycle in the Royal Roads Environmental Sciences Lab Manual (Royal Roads, 1998). The clover plant sample and the Azotobacter soil sample were provided with no details regarding sample location or collection.
The results section reports the observations. Interpretation of the results are discussed in more detail in the discussion section that follows the results section of this report.
Part A:
I. Rhizobium Isolation:
Gram Stain Cell Morphology:
The cell morphology of the Rhizobium cells from the crushed root nodule are summarised in Table 1.
Table 1. Cell Morphology of Rhizobium Cells from Freshly Crushed Nodule on Day 1.
| Cell Morphology of Rhizobium Bacteria Gram Stain | |
| Shape | rectangular rods |
| Axis | straight |
| Ends | rounded |
| Size Ratio (width:length) | 1:4 |
| Grouping | paired, chains, and clusters |
| Gram Stain | negative |
| Other | 3-5 round purple granules inside the rods |
The Rhizobium from the freshly crushed root nodule was Gram stained onto a slide and examined at 100X power with oil immersion. The bacteria cells were rods in chains, and clumps that stained Gram negative as indicated by the faint pink red colour of the rod membrane walls. Inside the rods, there were small rounded bodies or granules that stained a dark purple. There were three to five granules inside the rod cells. The cells are sketched below in Figure 1.
Figure 1. Gram Stain Drawing of Rhizobium Bacteria Cells from Crushed Nodule on Day 1.
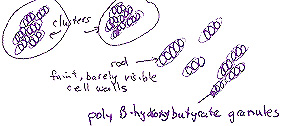
Colony Morphology:
The colony morphology of the Rhizobium from the crushed root nodule that was incubated on the Rhizobium agar plate at 30° C for 3 days is summarised in Table 2.
Table 2. Colony Morphology of Rhizobium .
| Morphology of Rhizobium Colony on Rhizobium Agar plate incubated at 30° C for 3 days | |
| Size | 2-4 mm diameter |
| Shape | circular |
| Pigmentation | colourless, same colour as the agar |
| Elevation | pulvinate, raised |
| Edge | smooth |
| Surface | smooth |
| Optical Characteristics | |
| Under Transmitted Light | translucent |
| Under Reflected Light | opalescent, glistening |
| Structure | amorphous |
| Consistency | viscous |
| Emulsifiability | n/a |
| Odor | musky |
| Other | large areas of run-on colonies that look like one large colony |
| Other Colonies | There were two other
colony types: Two large fuzzy round dark yellow colonies Many tiny small white round ones in linear patterns |
The plate was 50% covered with a solid mass of the Rhizobium spp. There were isolated colonies on the other half of the plate. In addition, two other types of colonies were observed.
Gram Stain Cell Morphology:
The cell morphology of the Rhizobium from an isolated colony described in Table 2 is summarised in Table 3.
Table 3. Cell Morphology of Rhizobium Cells from colony on Rhizobium Agar plate incubated at 30° C for 3 days.
| Cell Morphology of Rhizobium Bacteria Gram Stain | |
| Shape | rectangular rods |
| Axis | straight, bent or curved |
| Ends | rounded |
| Size Ratio (width:length) | 1:3 |
| Grouping | paired, chains, and dense clusters |
| Gram Stain | negative |
| Other | 3-5 round purple
granules inside the rods Some may be parts of broken rods Appear bloated and twisted |
The Rhizobium from the plate colony was Gram stained onto a slide and examined at 100X power with oil immersion. The bacteria cells were rods in chains, and dense clumps that stained Gram negative as indicated by the faint pink red colour of the rod membrane walls. Inside the rods, there were small rounded bodies or granules that stained a dark purple. There were three to five granules inside the rod cells. These rods were wider and more irregular in shape. Many of the rods were curved. The cells are sketched in Figure 2.
Figure 2. Gram Stain Drawing of Rhizobium Bacteria Cells from colony on Rhizobium Agar plate incubated at 30° C for 3 days.

II. Azotobacter Isolation:
Azotobacter Isolation in Flask:
A few granules of the Azotobacter soil was incubated in mannitol N-free medium for 7 days at 30° C. There were pellicles or opaque creamy light beige membranes like islands floating on the surface of the nitrate broth. The pellicles were irregular and round with diameters ranging from 2-5 cm. There was white fibrous needle-like growth on the upper air surface of the pellicles which was denser in the centre of the pellicles. There was a fine scum ring around the flask at the liquid-air interface. This scum was scraped and used to prepare the colony plate. The liquid was cloudy with yellow-brown fine solids in the bottom of the flask. In addition, there were two round brown fuzzy colonies growing in the center bottom area of the flask.
Colony Morphology:
The colony morphology of the Azotobacter from the incubated bacterium in the flask that was incubated on the mannitol N-free agar plate at 30° C for 4 days is summarised in Table 4.
Table 4. Colony Morphology of Azotobacter obtained from the Isolation in Flask.
| Morphology of Azotobacter Colony on mannitol N-free agar plate incubated at 30° C for 4 days | |
| Size | 1-5 mm diameter |
| Shape | circular |
| Pigmentation | creamy beige white, similar colour and hue as the agar |
| Elevation | pulvinate, raised |
| Edge | smooth |
| Surface | smooth |
| Optical Characteristics | |
| Under Transmitted Light | opaque |
| Under Reflected Light | opalescent, glistening, glossy |
| Structure | amorphous |
| Consistency | viscous |
| Emulsifiability | n/a |
| Odor | musky |
| Other | large areas of run-on colonies that look like one large colony |
| Other Colonies | There were no other colony types visible. |
The plate was 30% covered with a solid mass of the Azotobacter spp. There were isolated colonies on the other half of the plate. There were no other types of colonies were observed.
Gram Stain Cell Morphology:
The cell morphology of the Azotobacter obtained from an isolated colony described in Table 4 is summarised in Table 5.
Table 5. Cell Morphology of Azotobacter Cells from colony on Mannitol N-free Agar plate incubated at 30° C for 4 days.
| Cell Morphology of Azotobacter Bacteria Gram Stain | |
| Shape | rectangular rods, and mostly oval rods, many were peanut shaped |
| Axis | straight |
| Ends | rounded |
| Size Ratio (width:length) | 1:2 |
| Grouping | single, most were paired end to end |
| Gram Stain | negative |
| Other | dark outer membranes tiny purple dots inside the cell, decreasing density of dots closer to the center, but no clear and distinct vacuole in the center the cells looked like they were in different stages of division: some had narrower middles some had closely pinched middles some looked like two oval rods end to end |
The Azotobacter from the plate colony was Gram stained onto a slide and examined at 100X power with oil immersion. The bacteria cells were oval rods mostly in pairs, end to end, and many single cells. A relatively smaller quantity of short chains, and small clumps. The cells stained Gram negative as indicated by the faint pink red colour of the rod membrane walls. Inside the rods, there were very tiny dark purple dots that were denser closer to the rod cell membrane and the density decreased toward the center of the rod. Many of the cells appeared round and bloated. In addition, it appeared as the cells were in various stages of division, end to end. Many of the rods were peanut shaped. The cells are sketched in Figure 3.
Figure 3. Gram Stain Drawing of Azotobacter Bacteria Cells from colony on Mannitol N-free Agar plate incubated at 30° C for 4 days.

Part B:
Nitrate Reduction:
The test results for the detection of nitrite and nitrate in Durham tubes inoculated with four samples and a non-inoculated control are summarised in Table 6.
Table 6. Nitrate Reduction Tests on Bacteria in Nitrate Broth in Durham Tubes incubated at 37° C for 7 days.
| Sample Type |
Growth in Durham Tube |
Gas in Durham Tube |
Test for the presence of nitrite; tube colour after addition of reagents A , B |
Nitrite Test +=nitrite present; -=nitrite not present |
Test for the presence of nitrate; tube colour after addition of zinc |
Nitrate Test += nitrate present, not utilized -= nitrate not present, nitrate utilized |
| Pseudomonas aeruginosa |
Yes |
Yes |
Yellow No Change |
- |
Yellow No Change |
- |
| E. coli | Yes |
No |
Dark Red |
+ |
n/a |
n/a |
| Clover Soil | Yes |
Yes |
Yellow No Change |
- |
Yellow No Change |
- |
| Azotobacter Soil | Yes |
Yes |
Yellow No Change |
- |
Yellow No Change |
- |
| Control | No |
No |
Yellow |
- |
Dark Red |
+ |
Note: n/a indicates that this test was not completed due to the positive nitrite test result.
These results are interpreted in the discussion section.
The identification of the Rhizobium sp. is based on the cell morphology and the colony morphology. In addition, the growing media was selective to the Rhizobium sp. The Rhizobium agar is species specific with yeast extract to provide a source of organic nitrogen and mannitol as the carbon source in the growing media.
The cell morphology of the Rhizobium sp. is characterised as blunt rod shaped cells in pairs, chains and groups. They Gram stain negative and often appear banded due to the presence of poly b -hydroxybutyrate granules (Royal Roads, 1998). The Gram stains of the crushed nodule as described in Table 1 indicate the presence of bacterial cells that have the characteristic cell morphology of the Rhizobium sp. The rods were rectangular, straight and with blunt rounded ends and stained Gram negative. The rods were grouped in pairs, chains and clusters. In addition, they contained 3-5 small dark staining round granules that are characteristic of the poly b -hydroxybutyrate granules. The Gram stains of the isolated colony as described in Table 3 were very similar with the exception that they were thicker, and the shapes were more irregular. It appeared that some of the rods were broken. This may be due to the fact that the plates were refrigerated for four days after the incubation period prior to preparing the Gram stain. This means the colonies were older and could have deteriorated. In contrast, the root nodule Gram stain was of a freshly crushed specimen.
The colony morphology of the Rhizobium sp. tends to produce excessive amounts of slime, which is extracellular carbohydrate, when grown on media that is nitrogen limited and that contains carbohydrates, which would include mannitol. One colony plate was examined for the presence of Rhizobium sp. The colony morphology is described in Table 2. The most dominant type of colony was a large mass of white slime that covered more than 50% of the plate, with large patches of single colonies of the same morphology. In addition there were two other colony types that probably originated on the nodule, and are also capable of growth in this medium under the same incubation techniques. This does not exclude the possibility of contamination from other organisms in the soil, or the preparation technique.
The combination of the cell morphology of the Gram stains, the growth of the colonies on the Rhizobium selective media, and the colony morphology of the colonies provide sufficient evidence to demonstrate the successful isolation of a Rhizobium sp.
The most likely species of Rhizobium that has been isolated is Rhizobium leguminosarum bu. trifolii (R.l.bu.trifolii) which only associates with clover (Noble, 1998). This is a symbiotic and mutualistic relationship where both the R.l.bv.trifolii and the clover legume grow more favourably together than apart. The clover legume plant provides space, energy requirements and a low oxygen environment for the R.l.bv.trifolii. The Rhizobium provides fixed nitrogen for the plant. The process of host:bacteria recognition is chemically driven. The first stage is infection (Campbell, 1993). The roots of the clover plant secrete chemicals, including amino acids, dicarboxylic acids and flavonoid compounds, to attract R.l.bv.trifolii which are free living in the soil (Noble, 1998). The R.l.bv.trifolii move by chemotaxis to the root. The R.l.bv.trifolii bind to the root hairs which is mediated by lectins and causes the NodD protein of the R.l.bv.trifolii to activate the genes used in the formation of the nodules. The plant excretes tryptophan which is converted to indoleacetic acid which causes the root hairs to grow and curl around the R.l.bv.trifolii population (Atlas, 1998). Polygalacturonase is secreted by either the R.l.bv.trifolii or the plant and causes the root hair cell wall to soften and facilitates the entry of the R.l.bv.trifolii through the infection thread which is an invagination tube formed in the cell wall. At the end of the thread, the R.l.bv.trifolii are endocytosed . The plant cells proliferate in the cortex and the pericycle regions to form nodule tissue . The R.l.bv.trifolii develop into bacteriods and commence the process of nitrogen fixation. The vascular section of the plant is connected to the nodule. The plant provides sugar and organic substrates for metabolism; the bacteroids provide fixed nitrogen. The fixation of nitrogen depends on the nitrogenase enzyme system, which is very sensitive to oxygen. Rhizobium have unique problems in terms of energy production and the need for low oxygen tension to protect nitrogenase. Together, the plant and the R.l.bv.trifolii synthesise leghemoglobin which is a protein with an iron atom as part of its' prosthetic porphyrin ring that reversibly binds with oxygen. This molecule maintains a low oxygen environment in the root nodules by picking up the free oxygen and transporting the oxygen to the R.l.bv.trifolii for respiration to produce ATP used in nitrogen fixation (Campbell, 1993).
Under these experimental conditions it is inclusive to state that the Rhizobium sp. fixes atmospheric nitrogen because the Rhizobium sp. was grown in a medium that provided organic nitrogen in the form of the yeast extract. Rhizobium sp. can only fix atmospheric nitrogen under microaerophilic conditions, such as inside the root nodule (Royal Roads, 1998). It would be necessary to duplicate conditions found in nature in a strict artificial environment to prove that the Rhizobium sp was able to fix atmospheric nitrogen. Under artificial conditions, it would be essential to establish a closed system, and monitor the concentration of atmospheric nitrogen in the artificial environment in order to determine that Rhizobium sp. fix atmospheric nitrogen. This symbiotic relationship between the clover legume and the Rhizobium sp. enables the Rhizobium sp. to fix atmospheric nitrogen two to three times faster than nitrogen that is fixed by the Azotobacter sp., which is a free-living species in the soil.
The identification of the Azotobacter sp. is based on the cell morphology and the colony morphology. In addition, the growing media, mannitol N-free, is selective to facilitate the growth of the Azotobacter sp. The medium is nitrogen free, hence the Azotobacter must fix free atmospheric nitrogen, and mannitol is the carbon source. Consequently, those soil species of bacteria that cannot fix atmospheric nitrogen are selected against and only the species that can fix atmospheric nitrogen can grow in this medium.
Azotobacter sp. are aerobic bacteria and can grow at the liquid and air interface. There was significant growth of pellicles on the liquid surface and a ring of the same kind of film around the edge of the flask at the interface line. This area was scraped and grown on colony plates; an isolated colony was used for the Gram stain.
The cell morphology of the Azotobacter sp. is characterised as blunt rod shaped cells in pairs, chains and groups. They Gram stain negative and often appear banded due to the presence of poly b -hydroxybutyrate granules (Royal Roads, 1998). In addition, older cultures may contain cysts. The Gram stains of the cells obtained from an isolated colony as described in Table 5, indicate the presence of bacterial cells that have the characteristic cell morphology of the Azotobacter sp. The rods were rectangular, straight, with blunt rounded ends, and stained Gram negative. The individual cells were thick with a size ratio of width:length of 1:2. The rods were grouped in mostly in pairs, end-to-end, with some single cells. In addition, they contained tiny small dark staining round granules that are characteristic of the poly b -hydroxybutyrate granules. The density of the granules is higher closer to the cell membrane, and decreases towards the center of the cell. In older cells, there is often a characteristic vacuole in the center of the cell, which was not observed in this grouping of cells. The decreased density in the center of the cells could possibly be the initial stages of the formation of the characteristic cyst. These groups of cells appeared to be in various stages of dividing but it was impossible to tell as there was no growth and movement. There were long cells with small indentations in the center. There were other cells with a very distinct pinch in the center that formed peanut shapes. There were long cells similar to the peanut shape but the pinched area was closed. Depending on the thickness of the cell membrane at the pinch, these cells looked like one cell dividing into two cells or possibly a pair of cells, end-to-end.
The characteristic colony morphology of the Azotobacter sp. tends to be white, slimy, mucoid colonies. One colony plate was examined for the presence of Azotobacter sp. The colony morphology is described in Table 4. The only type of visible colony was a large mass of white slime that covered 30% of the plate, with large areas of run-on colonies and some single colonies of the same morphology. The growing media of the mannitol N-free agar plate is selective in that it is nitrogen free and uses mannitol as the carbon source. It supports the growth of organisms that can fix free atmospheric nitrogen and use mannitol as the sole carbon source.
The combination of the cell morphology of the Gram stains, the growth of the colonies on the Azotobacter selective media, and the colony morphology of the colonies provide sufficient evidence to demonstrate the successful isolation of an Azotobacter sp. While this Gram stain did not have distinctly visible cysts, it is possible that the cysts would have been visible if the colony had been one or two more days older.
The Azotobacter sp. is an aerobic organism that can fix atmospheric nitrogen as demonstrated by the grown in the flask at the liquid/air interface. The presence of oxygen can deactivate the nitrogenase enzyme and inhibit the fixation of nitrogen. The Azotobacter sp. compensates for this problem by maintaining a very high respiration rate which effectively uses the oxygen as soon as it enter the cell.
The process of nitrification occurs when ammonia (NH4+ )or ammonium ions are oxidized to nitrite ions (NO2- ) , and then to nitrate ions (NO3-). Each step in the process is facilitated by a different type of organism.
The first step of oxidising the ammonia to nitrite can be facilitated by heterotrophic Arthrobacter or autotrophic Nitrosomonas (Atlas, 1998). This oxidation reaction is:
Equation 3. Nitrification: Oxidation of ammonium to nitrite.
The second step of oxidising the nitrite to nitrate can be facilitated by heterotrophic bacteria such as the Arthrobacter, Aspergillus, and by autotrophic bacteria such as the Nitrobacter. This oxidation reaction is:
Equation 4. Nitrification: Oxidation of nitrite to nitrate.
The two step process produces energy which is used to assimilate CO2. The significance of nitrification is that it converts the positive charge on the ammonia ion to a negative charge on the nitrate and nitrite ions. This effectively polarises the molecules, makes them soluble in water, and consequently more mobile in the environment. The soluble form can be taken up by plant roots, and used for assimilation into organic compounds. Any significant stresses on the soil environment can reduce the presence of nitrifying bacteria. In addition, significant leaching can remove the soluble form of the nitrogenous ions, and reduce the fertility of the soil.
Nitrate reduction is the process of reducing nitrate ions to various nitrogenous products (Atlas, 1998). Two types of nitrate reduction include assimilatory and dissimilatory. Assimilatory nitrate reduction uses enzyme systems, including nitrate and nitrite reductases, to form ammonia which can be assimilated into amino acids and proteins. This process is occurs when there is oxygen. Conversely, under anaerobic conditions, dissimilatory nitrate reduction occurs and nitrates are converted to reduced products, and organic matter is oxidized. There are two types of dissimilatory nitrate reduction. In some cases, facultative anaerobes reduce nitrate to nitrite; in other cases the nitrite is reduced to nitrogen gas.
Denitrification is the process that converts nitrates to gaseous nitrogen (N2) in a multi-step reaction (Atlas, 1998). The reaction is:
Equation 5. Denitrification Multi-step reaction.
This process occurs mostly under anaerobic conditions. The most dominant denitrifying bacteria include the Pseudomonas and the Alcaligenes, but also include the Azospirillum and the Rhizobium.
The experimental results for the nitrate reduction tests are summarised in Table 6. Five Durham tubes of nitrate broth medium were used to test different samples, including a control tube that was not inoculated. After the inoculation period, the medium was reacted with two reagents, A and B, to test for the presence of nitrate and nitrite. Reagent A, sulfanilic acid, and reagent B, an aromatic amine, react with nitrite to produce a dark red azo dye product. If the tube tested negative for the presence of nitrite, then a zinc catalyst was introduced to test for the presence of nitrate. The zinc catalyst reduces nitrate to nitrite, which then proceeds in the previous reaction to produce the red azo dye. If the tube is cloudy this indicates the presence of bacterial growth. If there is a bubble in the base of the tube, this indicates the presence of a gas.
The test tube inoculated with Pseudomonas aeruginosa had both growth and gas in the tube. It did not test positive for either nitrite or nitrate, which means that the organism converted the nitrate in the broth to nitrite to nitrogen gas. This is characteristic of the Pseudomonas which is a denitrifying bacteria. The evidence supports this observation.
The Durham tube inoculated with E.coli had growth and did not have gas in the tube. This tube tested positive for nitrite. This organisms is able to convert nitrate in the broth to nitrite. It was not possible to test for the presence of nitrate in the broth. It cannot convert nitrate to nitrogen gas as there was not gas in the tube.
The Durham tube inoculated with clover soil had both growth and gas in the tube. It did not test positive for either nitrite or nitrate, therefore the nitrate was utilised by the organisms in the soil and converted to nitrogen gas by denitrification. The microbe associated with clover soil is the Rhizobium which is characteristically known as a nitrogen fixer. The logical assumption is that the Rhizobium in the clover soil is responsible for the denitrification of the nitrate in the broth medium and converting the nitrate into nitrogen gas as evidenced in the tube.
The Durham tube inoculated with Azotobacter soil had both growth and gas in the tube. It did not test positive for either nitrite or nitrate, and therefore the nitrate was utilized by the organisms in the soil and converted to nitrogen gas by denitrification. This organism is capable of denitrification.
The Durham tube inoculated with nothing represented the control tube. There was no visible growth or gas in the tube. The control was negative for nitrite and nitrate. This means that there was no contamination in the media or reagents used in this procedure that would cause a reaction to proceed. This increases the surety of the previous results in that those results may be attributed to the nature of the inoculating substance and not to the materials or the procedure used.
In conclusion, it is the combination of the growth of organisms on selective medium, the cell morphology of the Gram stains, the colony morphology of the agar colony plates, and the nitrate reduction tests that provide the evidence to conclude the following.
The Rhizobium sp. was isolated from the root nodule of the clover legume. Rhizobium sp. is capable of denitrification. While this evidence was inconclusive in the colony plates, the evidence is clear in the nitrate reduction tests in the Durham tubes.
The Azotobacter sp. was isolated from the Azotobacter soil sample. This organism is capable of fixation of atmospheric nitrogen as evidenced by the growth of the isolated culture in the medium that did not contain any form of nitrogen. In addition, this organism is capable of denitrification as evidenced by the absence of nitrite and nitrite in the nitrate reduction test.
Atlas, R. and Bartha, R. 1998. Microbial Ecology:
Fundamentals and Applications. Fourth Ed
Benjamin Cummings. Menlo Park, Ca. 694 pp.
Campbell, N. 1993. Biology. Third Ed. Benjamin Cummings. Redwood City, Ca. 1190 pp.
Noble, M. 1998. Lecture Notes. "Nitrogen Fixation
in Root Nodules of Legumes".
Friday, February 6, 1998. Royal Roads University.
Victoria, B.C.
Royal Roads Environmental Science Lab Manual. 1997.
Experiment 12: Aspects of the Nitrogen Cycle. pp. 12-1 to
12-12.
Royal Roads University. Victoria, B.C.
Please be aware that this is a student level lab and has not been peer reviewed by a professional journal. Please refer to primary journal publications for additional information for your research.
Student labs are my favourite part of learning because each is a prototype mini-project with multiple stages in: planning, doing, analysing, and writing.

 email Waterose
email Waterose
Please Sign My Guestbook
Please View My Guestbook

| Articles | Projects | Resume | Cartoons | Windsurfing | Paintings | Album |
 | ||||||