Namely, in both types of insects, engrailed is expressed in a pattern of regularly spaced stripes, one stripe per segment. However, in Drosophila, engrailed is thought to be directly regulated by the primary pair-rule gene even-skipped, while in the grasshopper the eve homologue does not serve a pair-rule function in early development (Patel et al., 1992). In Drosophila, the eve gene is required for expression of all 14 engrailed stripes. Early eve expression acting as a bell-shaped concentration-dependent morphogenetic field, biased by combinatorial interactions with other primary pair-rule genes, subdivides each of its domains into multiple functional subregions.
In more advanced insects, like the beetle Tribolium, intermediate segmentation control system seems to be present (Sommer & Tautz, 1993; Wolff et al., 1995). The gap gene hunchback (hb) plays a central role in determining the anterior-posterior pattern in the Drosophila embryo. The HB-product morphogenetic gradient may have the same functions in early Drosophila and Tribolium development, despite the different types of embryogenesis in these two species (long versus short germ development). In Drosophila the HB gradient activates Kruppel which plays a key role in determination of the thoracic and abdominal segments. Most importantly, Tribolium hb could be involved in Tribolium Kruppel regulation in a similar manner to that in Drosophila, namely by repressing Kruppel at high concentrations and activating it at lower concentrations (Schulz & Tautz, 1994).
In Drosophila, the third and the fourth primary pair-rule gene hairy stripes lie within the region of the Kruppel expression domain. Analysis of Kruppel expression indicates that the same is true for Tribolium. The first expression of Kruppel precedes the expression of hairy in the same region. The initially single domain of Tribolium hairy expression then splits into two. It is known that hairy stripes three and four form at first a single domain in Drosophila as well, which later splits into two stripes.
The striking parallels in the gene segmentation mechanisms between Drosophila and Tribolium suggest that the patterning mechanism through a transient pair-rule pre-pattern is a fundamental component of the segmentation process in insects. However, not all pair-rule genes are involved in the pair-rule prepattern in the case of more ancient insect embryogenesis.
As Jäckle with co-workers noted, currently nobody knows whether the multiple regulatory circuits discussed above represent a fine tuning system to specify the precise position in the embryo where the target gene is active, whether the regulatory inputs are redundant, or whether some of the regulatory pathways represent just evolutionary relics which are not decisive in embryo development (Jäckle et al., 1992).
Bringing to mind my computational results, one could suggest that the evolutionary growth of the pair-rule and the segment-polarity networks has features prescribed by my model. However, my scheme is apparently applicable only for the thoracic and abdominal region segmentation mechanisms (in the vicinity of the Kruppel band of expression). In particular, the engrailed activation cascade apparently recruited new pair-rule type genes, even-skipped, first of all, on the evolutionary path from the short to the long germ-band embryogenesis (figure 11).
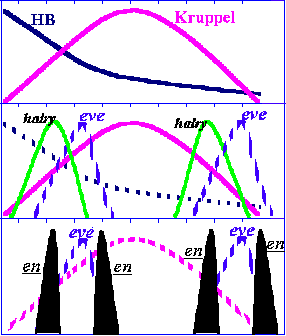
Figure 11. The HUNCHBACK morphogenetic gradient activates, in a concentration-dependent manner, the Kruppel gene. In turn, the bell-shaped morphogenetic gradient of KRUPPEL activates two stripes of the primary pair-rule gene hairy and alters the activity of the even-skipped gene. Finally, the bell-shaped morphogenetic fields of the EVE activate pairs of the engrailed gene expression stripes. This finding of Fujioka et al. (1995) adds to the previous discoveries of embryo-length morphogenetic gradients, composed of maternal gene products and the more localised gradients of gap gene products, and shows that Drosophila uses the same principle again to further refine pattern information.
Another prospective application of my scheme, I believe, is to the evolutionary appearance of highly symmetrical polyradiate patterns of the segmentation genes in Drosophila imaginal discs.
My computational experiments allow an evolutionary appearance of the Drosophila segmentation mechanism to be simulated and tested. I suggest that the evolutionarily fast formation of the fly segmentation scheme is facilitated by self-enlargement of the initial segmentation cascade via recruiting of new gene members.
Our computer experiments were created as a step towards the long-range goal of using simulation as a tool to study of evolution of real gene networks. The inspiration from our results seems to consist in a fresh interpretation of gene mechanisms for macroevolutionary steps. According to our interpretation of the computational findings, material for evolutionary selection is represented not by bearers of unique prospective mutations but by bearers of prospective self-enlarged gene networks/cascades. Apparently it is impossible get over enormous gaps between different levels of organization by means of little steps of unique nucleotide substitutions. Evolutionary changes of body plan apparently require the co-ordinated change of several phenotypic characters to produce adaptive variants. But this requires simultaneous mutations at several genes. Self-assembled networks ensure a mechanism for stepped evolutionary growth of gene regulatory ensembles, while coordinately changing several genotype-phenotype characters. The self-assembled networks give evolution a chance to overcome the gaps between different levels of organisation.