


Host genome initially consists of functionally coupled pair of gene regulatory elements (O-gene + A-gene).

"Maternal" gradient of primary morphogen (like Drosophila's bicoid morphogen) is predetermined. The exponential-in-distance morphogen gradient activates O-gene in concentration dependent manner. In its turn, O-product will activate A-gene in concentration-dependent manner also. Concentration profiles of the O- and A-products for wild species have following simple view:
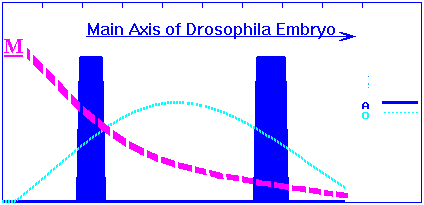
and corresponds early embryo phenotype with two bands of the A-gene expression:

The wild-type genomes are two-string variables. Namely, the A-gene string is
CATAATnCATAATnCATAATn,
where A, T, G, C is four-letter DNA code and n is spacer. CATAAT belongs to targeting sequences for O-product binding. The O-gene string has similar view.
For the purpose of oversimplified but indirect implementation of the driving force, I assume appearance of a "virus". The virus randomly transmitted from "carriers" to health genomes. By definition, the virus successfully transmitted if host genome has in the A-gene O-binding site. I assume it inserts in the A-gene by cutting of the O-binding site and becomes silent. With predetermined probability the virus wakes up and with time gradually decrease host's reproductive potential, finally killing the host, thus eliminating affected genome from evolution.
Point mutations in the O-binding sites lead to insensitivity to virus. However, "wild" type of our genome could not lack the O-binding sites in A-gene, because of consequent lacking of normal phenotype (the A-product concentration profile). Hence, in case of genome with absence of the wild-type O-binding site in the A-gene (but with normal pattern of A-gene expression), this perspective mutant will be insensitive to the virus and obtain selective advantage. In time, the mutant genome will exclude wild type. This is, of course, a model example of host-parasite evolution.
By the way, this reminds well-known case of human crescent-cell anemia. Namely, bearers of the mutant hemoglobin gene suffer from anemia, but resistant to malaria. As a result, selection leads to spreading of the mutant gene among native inhabitants of Africa.
I must emphasize that such selection by parasite pressure is effective if only the design of the wild genome allows appropriate reorganisations in principle. Virus acts as a catalyst of the process. It does not produce but catch up new forms. If testable in calculations wild-type genome have not appropriate potential for reorganization, a virus will not help. Then, I imply broad interpretation of "virus", that is it may be plasmid or another genome element.
I assume also that the virus sequence specificity can mutate (with very low predetermined probability). As a result the mutant virus will can find and insert in other site then wild-type O-binding one. But the mutant still can insert only in A-gene or its extra-copies and offspring genes.
See also: Ikegami T. and K.Kaneko, 1992, Evolution of host-parasitoid network through homeochaotic dynamics, Chaos, 2, 397-408
Kaneko K. and T.Ikegami, 1992, Homeochaos: dynamics stability of a symbiotic network with population dynamics and evolving mutation rates, Physica D, 56, 406-429
Ikegami and Kaneko have studied a population dynamics model with interaction among hosts and parasites, mutation and mutation of mutation rates. Each species is coded by a string and the interaction between host and parasite depends upon Hamming distance between their strings. According to authors, when the host-parasite interaction is strong, mutation rates are sustained at a high level, where many species form a network. The network consists of species connected by single point mutations and many species are percolated in the gene space.
This somewhat remainds my simulation results.