IntroductionWARNING: heavy-going reading ahead. Don't read this page unless you feel compelled by curiosity. You can probably answer all the queries you have about e-den syntax simply by playing with the Bug Lab ... And besides, you don't need to know any e-den syntax to play host to your share of the e-den master grid. Still here? Don't blame me then.
The interpretation of each digit of the e-den genomes is at all times guided by two parameters:
the gene mode and the gene sub-mode. There are 11 gene modes, one for each digit
plus "Junk" mode (gene mode 10). (The major gene modes and the sub-modes applicable to each
are explained in detail below). The zero gene mode is not being used, at present,
and defaults to Junk mode. (In fact, earlier descriptions of the e-den syntax treated them as
synonymous). In designing your organisms, however, this default action should not be relied
upon as it is likely to change.
Genes and Non-genes:Junk ModeThe genomes of e-den have sections that are meaningful and contribute to the structure of the organism - the genes - as well as sections of "junk" that separate the meaningful segments. These junk sections were added, in part, to prevent frame-shift mutations; that is, disruptions of the syntax downstream from a single-point deletion or insertion mutation. Because of these junk sections, the syntax of each gene is independent of the others and holds its meaning provided its start and end boundaries are left intact. These boundaries are usually marked with a nine. Thus, a typical gene begins with a nine, followed by a digit defining the gene mode, a variable stretch of other digits and then, to end the gene, another nine. Once a gene has ended, what follows is considered junk until a new gene is started by a 9n combination, where n represents a digit from 1 to 8.For the gene modes 1 to 8, the digit used to represent the gene mode within the syntax-interpreting function is the same as the actual digit that introduces the gene. Thus, Skeletal genes begin with a 1, Visual genes with a 2, and so on. In the following string, the junk is indicated with square brackets and a visual gene by parentheses; the spaces are only provided for readability. ....) 9 [123456787654321] 9 (2501) 9 [123456787654321] 9 (..... Note that a zero was not necessary to introduce the sections of junk in this example and, although the combination 90 is currently interpreted as the start of a junk sequence, this is not the recommended approach. In fact, the combination 90 should be avoided. If you have used the Bug Lab, you might have noticed that junk sections consisting of 9s are often inserted between genes. The number of consecutive 9s in such a section is irrelevent. Although the 9 digit is best thought of as the "New Gene" marker, a string of 9s leaves the syntax poised at the start of a gene without actually leaving Junk mode. The junk section only ends when one of the nines is followed by a digit 1 to 8. Thus, a junk section can consist of any number of non-nine digits, followed by any number of consecutive nines but then ends with the first non-nine digit after that. Within each gene, a 9 is required to end the gene and to return to Junk mode but a 9 only has that effect if the gene sub-mode is "On StandBy". With other sub-modes, 9s can have a variety of meanings including the integer 9 itself, when this is being assigned to one of the organism's internal parameters. Thus, although 9s represent the major "punctuation mark" of the e-den genomes, it is not possible to blindly scan a genome and separate the Junk from the non-Junk merely by noting the positions of the 9s. Each digit must be interpreted sequentially.
The Major Gene ModesWithin each gene mode, the syntax is slightly different although there are common themes. Typically, each gene begins with the default sub-mode, "On StandBy", and the first digit within the gene sets a new sub-mode. For some sub-modes, this has an immediate effect (such as calling a function within the embryological software) and the sub-mode then reverts to "On StandBy" before the next digit is read. For other sub-modes, entering the sub-mode indicates that a particular parameter is about to be assigned a value and the following digits are used to assign the value - the sub-mode does not revert to "On StandBy until the expected number of digits has been read. Rarely, a sub-mode offers new choices of sub-sub-modes.In the Bug Lab, the following convention is used beside each digit: a forward arrow indicates that choosing this digit will take you deeper into the syntactical tree or, at least, take you to a new window where you will asked to provide the necessary number of digits to assign to the value you have chosen; a backwards arrow indicates that choosing this digit will take you back to a more basic syntactical level or a previous screen; a colon (:) indicates that the sub-mode will revert immediately to the current one so that the screen will not change. If you have not already played with the Bug Lab to see the effect of choosing various digit combinations, it is recommended that you do so now. In fact, it is a good idea to print this page and have the Bug Lab open as you read it. When a genome is being analysed, the initial gene mode is always "Junk". After the first 9 occurs, indicating the start of a new gene, the next non-nine digit sets one of the major gene modes as described in more detail below:
0 Reserved Reserved ModeMajor Gene Modes... Top... Contents... The Skeletal ModeThe Skeletal Mode has the simplest syntax of all modes. Only the first digit of each skeletal gene (after the obligatory 91) has any effect. The genes 91x9 and 91xyyyyyyyyyyyyyyyy9 (where x is a number from 1 to 8 and yyy is any sequence of non-nine digits) both have the same effect and simply append the digit x to the organism's skeleton. In the Bug Lab, these options are labelled: 1 skin, 2 eye, 3 digestive, 4 spike, 5 signal, 6 armour, 7 right turn, 8 left turn. (For some of these, the situation is more complex and the 1 segment, in particular, might be better thought of as an ear. The function of each body segment was reviewed in the e-den biology page).Only a zero at the start of the gene (as in 910...) leads to new syntactical level; the sub-mode is changed to "skeletal type" and the next digit sets a parameter ("skeltype") that determines the way joints move under various tensions. This is a relatively recent addition to e-den biology and is not explained in detail anywhere in this site but the names in the Bug Lab are fairly self explanatory (mobile vs frozen). After the skeletal type has been set, all further digits are ignored. Thus 910x9 and 910xyyyyyyyyyy9 are equivalent. Within the Bug Lab, the ignored digits are referred to as Junk but this is in some ways misleading; the gene mode remains Skeletal and the sub-mode is "Finished/Ignore". As with most gene modes, a 9 ends the gene and causes the gene mode to revert to Junk. Major Gene Modes... Top... Contents... The Visual ModeThe Visual mode and General Neuron mode have an almost identical syntax. The Visual mode is used for setting the properties of several visual neurons at once, however, whereas the General Neuron mode only sets the properties of a single neuron at a time. A typical Visual gene begins with the digits 92x whereas a typical General Neuron gene begins with 97x. In both, the initial sub-mode is "On StandBy" and the subsequent sub-mode is determined by x. Unless x equals 9 (which ends the gene), a number of digits are read following the x and assigned to the parameter indicated. The number of digits read depends on the sub-mode and ranges from 1 digit, in the case of "Drift", to 7 digits in the case of "Axon" sub-mode or "Dendrite" sub-mode. Following these digits, the sub-mode reverts to "On StandBy" and the whole cycle can be repeated indefinitely until a 9 occurs while the sub-mode is "On StandBy". (Don't worry if this seems confusing as an example will follow).The available sub-modes are shown below, with the number in square brackets indicating the number of digits read within each sub-mode:
0 Restore Tendency [2] A Visual Gene such as ...925121294519... put through the decoding utility (decode.exe) might look as follows: junk, junk, junk.....9
(visual) junk,junk,junk....
This gene sets the "current visual neuron" to number 12, assigns the value 29 to
that neuron's threshold, assigns the value 51 to that neuron's sensitivity and then ends.
Any number of extra lines after the 2(start) and before the 9(end) could be inserted,
provided they were of the form junk, junk, junk.....9
(visual) junk,junk,junk.... The above sequence would instead be interpreted as: junk, junk, junk.....9
(visual) Note that this means that single-digit neuron numbers (1,2,3 etc) must be referenced as 01,02,03 etc in Visual mode, or as 001, 002, 003 etc in General Neuron mode. For most sub-modes, the digits supplied after the sub-mode is entered are interpreted literally as a 2 or 3 digit integer. For the drift parameter, the equation drift=2n-9 is employed, so that drift is assigned avalue ranging from 9, if n equals 9, to -9, if n equals 0. For the axon parameter, a seven-digit string is used to assign values to more than one parameter. The first digit is used to choose an axon number from the 10 available (0-9); the second digit is used to select an axon type (there are arbitrary associations between each digit and each type); the next three digits determine the recipient of the axon, the neuron that will receive its ouput (note that this is a three digit number for both visual and general neuron genes); the final two digits determine the effect of the axon, which corresponds to the strength of the output or the synaptic weight. A similar seven digit sequence is used for specifying dendrites. WARNING: There is a potential trap here for the unwary. The zero-numbered axon and dendrite do not actually exist as axons and dendrites. They provide a method of allowing neurons to modify themselves and can be used to encode long term potentiation, for instance. Regardless of what recipient is specified for the zero-axon or dendrite, the software redirects the axon or dendrite to its own neuron. Although this conjurs up an image of an axon leaving its own neuron and looping back, this makes no biological sense and it would be better to think of these cennections as fictions standing in for some intrinsic cell process that self-adjusts the neuron. The software, however, treats the axon and dendrites as though they did loop back on to their source neuron.
The 25 prototype visual neurons can be considered as a prototype visual segment. Each of these prototype neurons is actually a set of neurons, taken from all the visual stacks, such that each member of the set occupies the same segment position. This segment position is the same as the visual neuron number. Thus, in a one-eyed organism, prototype visual neuron #01 is the set of neurons #101, #126, #151 and #176. In a two-eyed organism, the same prototype is the set of neurons #101, #126, #151, #176, #201, #226, #251 and #276. If you examine the vision section on the neurology page, you will see that each these neurons receives information about the distance to the nearest one-atom (grass-atom). Thus, while neuron #101 is the neuron that receives this information from the east visual field of the first eye, prototype visual neuron #01 is the set of all grass-seeing neurons, including #101 and also those with neuron numbers 101+25n, for a range of values of n. Similarly, #02 is the set of all two-seeing neurons, #03 is the set of all three-seeing neurons and so on. The visual genes allow you to change all of these functionally similar neurons at once, rather than repeating genetic instructions for each of the neurons in the set.
By convention, when specifying neural connections for these prototype neurons, they are considered to be equivalent to the first segment of actual visual neurons, the real neurons #101-#125; these neurons
process information from the east visual field of the organism's first eye. (For more details on the arrangment
of neurons processing visual information, consult the
e-den neurology page ).
If (and only if)
the organism has already specified that its skeleton contains an eye, assigning
a value to visual neuron #12 automatically assigns the value to neuron #112. An
appropriate value is also assigned to the similarly positioned neurons in the south, west
and north facing neuronal segments, in this case #137 (112+25), #162 (112+2x25) and
#187 (112+3x25). If a second eye has already been appended to the developing organism,
the same procedure is applied to neurons #212,#237,#262,#287, and so on until the organism
runs out of eyes or neurons.
Not every gene needs to specify which neuron is to receive the assigned values. The current neuron number and current visual neuron number
retain their values across genes. One gene can set the current visual neuron number to #12
(....925129.....) and a later gene can assign a value to its threshold (.....921299.....). Major Gene Modes... Top... Contents... The Metabolic ModeThis mode has a relatively simple syntax. Each metabolic gene begins with 93 and ends with 9, with a variable number of assignment statements in between. Each assignment statement has the form x(sub-mode)nnn, where x is a digit from 1 to 8 that specifies which parameter is about to be adjusted and nnn is a sequence of either 2 or 3 digits depending on the actual sub-mode, as shown below:
0 Not in use [0]
The 3-digit values assigned to Metab-Div and Metab-Child are first multiplied by 10 and thus these parameters
can have values ranging from 0 to 9990. The other parameters have values ranging from 0 to 99.
It is important to note that the first assignment to each of these parameters is interpreted literally but,
thereafter, the assignment contributes to a rolling average that has a 50% contribution from the new value
and 50% from the previous average. Major Gene Modes... Top... Contents... The Change Current Neuron ModeThis mode is not supplied for the benefit of the human programmer but rather for the evolutionary process itself. Its major effect is to change the value of the current neuron number, thereby changing the target of assignment statements in later General Neuron genes. For the human programmer, it is always easier to use the General Neuron mode itself:......9
(general neuron)
Although the above statement could mutate into statements about another neuron if any of the
digits "112" mutated, neuronal genes that failed to specify a neuron number and simply
relied on the previous value could not mutate in this way. And, when a mutation occurred,
all assigment statements referring to that neuron would be re-assigned as a result; there
would be little chance for evolution to keep some of the assignment statements with the original neuron
and move others to a new neuron. It could happen, if the combination 975nnn occurred at the right
point in the genome, but this is relatively unlikely. The actual operation of this syntax is largely self explanatory, with the following options being available:
0 Reduce current number by 100
Genes in this mode are of the form 94x... and when x is 0-5 the current neuron number is changed as
indicated and the sub-mode reverts to "On StandBy", offering the same choices again. When x is 6, a new
syntactical level is reached and a single digit is read before returning to "On StandBy"; that digit causes neural
parameters to be copied from the current neuron to one of its neighbours or vice versa, or to the "generic" neuron
or vice versa (the generic neuron is number #000, the master prototype). Major Gene Modes... Top... Contents... The Multiconnect ModeThis gene mode was designed with both the human programmer and evolution in mind. If you create an organism with a random genome in the Bug Lab, insert it in the Close-Up window and then select it by right-clicking it, you can look at its neural connections in the Net Window. The following neural structure was created with a purely random genome about 1000 digits long: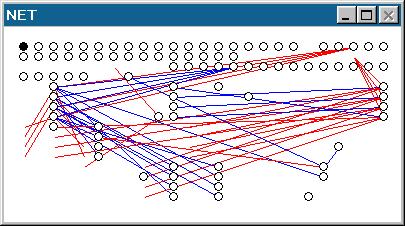 The syntax of this gene mode is relatively complex. Starting in the "On StandBy" sub-mode, the following options are offered (the number in square brackets represents the number of digits expected after the sub-mode is set):
0 Set recipient to current [0] Sub-modes 1 to 7 are used to assign values to the various parameters required by the Multiconnect() function; in each case the necessary number of digits is read and then the sub-mode reverts to "On StandBy". There are 8 parameters required for the function: start, end, segments, connection rule, recipient, axon number, axon type and effect. The start and end parameters are set by the six digits read in the "Range" sub-mode. These two parameters set the range of neurons that will be susceptible to the Multiconnect() function. The start parameter has a default initial value of #101, which represents the first visual neuron, and the end parameter has a default initial value equal to the maximum neuron number, currently #400. Thus, if these parameters are not adjusted, the Multiconnect() function applies to all neurons after the first stack of 100. (Note that these parameters are shared with the Neurocopy() function and can also be adjusted in the Neurocopy sub-mode; they retain their values across genes). The segments parameter further limits the range of neurons susceptible to the Multiconnect() process. It refers to which segments of 25 neurons within each stack of 100 will have connections applied. If the parameter has a value of 1,2,3 or 4, then the 1st, 2nd 3rd or 4th segments respectively of each stack will be susceptible. If it has a value of 5, then the 1st and 2nd segments will be suceptible, if 6 then the 3rd and 4th segments, if 7 the 1st and 3rd, and, if 8, the 2nd and 4th. Values of 9 or 0 leave all segments within the start-end limits susceptible. The neurons indicated by the combination of the range and segments sub-modes are those that will develop new axons during application of the Multiconect process. There are four possible connection rules, which are assigned with the genetic sequence 3n; the rule chosen depends whether n takes one of the values indicated in square brackets: FORWARDS [0,1,2], BACKWARDS [3,4,5], SPECIFIC [6,7,8] and EQUIVALENT [9].
The recipient parameter is a 3-digit number that is used to determine where the new axons are directed.
In the case of the SPECIFIC connection rule, the recipient parameter indicates the number of the neuron to
which all new axons will be directed. In the case of the FORWARDS or BACKWARDS rules, this parameter indicates
the interval between the axonal source and axonal target; if the connection rule is FORWARDS and the interval is 111
then all neurons satisfying the range/segments restrictions will be connected to a neuron 111 positions further
along in the neuronal array (provided such a neuron exists). If the connection rule is EQUIVALENT
then some simple directional logic is applied, similar to that employed in the
visual gene mode (see above).
This software interprets the recipient parameter as a literal recipient number for the first neuron in the range,
and then tries to find a functionally equivalent recipient for every other neuron in the range. The final parameters needed by the Multiconnect() function are the axon number, axon type and axon effect for the new connections. To continue our previous example, the entire set of parameters might specify that for all neurons in the range #101-#400 that fall within the 1st and 3rd segments of their respective stacks, their fifth axon should be of type 1 (direct excitatory) with an effect of 25, and should be connected to a neuron 111 neurons further along in the array (start = 101, end = 400, segments = 7/ODD, rule = 1/FORWARDS, recipient/interval = 111, axon number = 5, axon type = 1/EXCITATORY, effect = 25). Note that these parameters may be set in any order and need not be re-specified with each Multiconnect gene; they retain their values across genes. Also, note that the Multiconnect() function is not actually called unless the digit 8 is read while in "On StandBy" mode. It is possible to end the gene (with a 9) without calling the function at all. Finally, when a zero occurs in "On StandBy" mode, it has two effects: it assigns the value of the current neuron number to the recipient parameter and also sets the connection rule. No digits are read and the sub-mode remains "On StandBy". Major Gene Modes... Top... Contents... The Alphabetical ModeThis simple mode sets the species designator or name of the organism. Basically, in "On StandBy" mode, it ignores all digits >2 (except 9, which ends the gene). Each occurrence of 0,1 or 2, however, is regarded as the first of a pair of digits; the second digit is read and then the sub-mode reverts to "On StandBy". This gene mode, then, collects two-digit numbers ranging from 00 to 29. Zero-zero resets the position counter to its default starting position; the other numbers stand for letters (01 to 26 for a to z) or punctuation (27 for space and 28 for hyphen). Each new character is used to fill in an array which may be up to 20 characters long. This allows users to name or identify species and is also used, in part, to determine whether two organisms can reproduce sexually.Major Gene Modes... Top... Contents... The General Neuron ModeThis gene mode has an identical syntax to the Visual gene mode and is discussed above.Major Gene Modes... Top... Contents... The Neurocopy ModeLike the Multiconnect mode, this gene mode allows either the human programmer or evolutionary processes to specify multiple neuronal features at once by calling a specific function. Like the Multiconnect() function, the Neurocopy() function takes a number of parameters that can be set within this gene mode and these parameters retain their value across genes. Unlike the Multiconnect mode, however, ending a Neurocopy gene automatically calls the Neurocopy function.The syntax of this gene mode is complex and is best explored using the Bug Lab, which offers the following options:
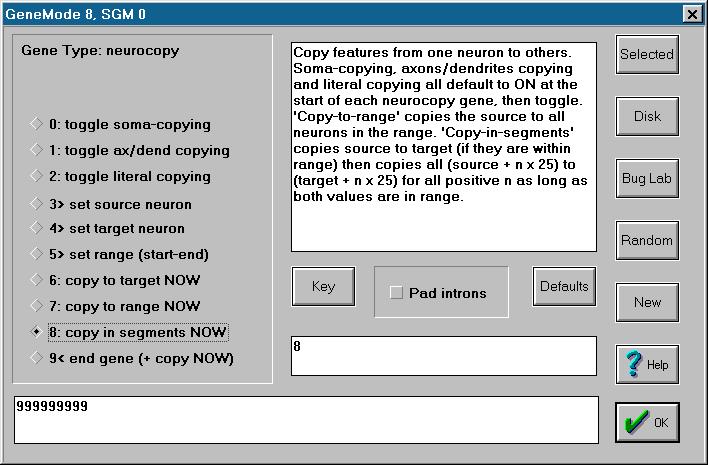 The major parameters required by the Neurocopy function are the source, target, start and end parameters, with the start and end parameters being set by six digits set within the Range sub-mode and the other parameters set by 3 digits within their own sub-modes. Note that the start and end parameters are shared with the Multiconnect mode. In addition, the function needs three Boolean (TRUE/FALSE) parameters: copy soma, copy axons and dendrites, and copy literally. These all default to TRUE or ON with each new Neurocopy gene and thereafter toggle with each occurrence of the relevant digit (0,1 or 2). Finally, the function can be called with one of three copying rules: source-to-target, source-to-range, or copy-in-segments. Each occurrence of the relevant digit (6,7 or 8) immediately calls the function with that rule, using whatever values the necessary parameters hold at that time. A 9 both calls the function with the source-to-target rule and also ends the gene. The function uses the supplied parameters as follows. Basically, it copies parameters from the source neuron to one or more target neurons. If copy-soma is ON, then the values of threshold, drift, flux, sensitivity, restore-tendency, and post-fire-change are copied (these are collectively know as the "soma" variables). If copy-axons-and-dendrites is ON, then the neural connections are copied, overwriting previous uses of the relevant axons and dendrites (but not deleting existing connections in the target neuron if none are specified for that axon or dendrite in the source neuron). If copy-literal is ON, then the soma variables simply overwrite the previous values in the target neuron. If copy-literal is OFF, however, then the source values are averaged with the target values to create a neuron with characteristics intermediate between the two. For connection variables, the copy-literal parameter determines whether the source connections are copied blindly, so that the target neuron ends up connected to exactly the same neurons as the source neuron, or whether the target neuron is provided with a functionally equivalent connection, in the sense that was discussed under the Visual gene mode (see above). The three copying rules determine which neurons are the target of the copying process. The source-to-target rule simply copies values from the source neuron to the single neuron specified by the target parameter. The source-to-range rule copies from the source neuron to every neuron in the range specified. The copy-in-segments rule copies the source neuron to the target neuron, adds 25 to both and the re-copies, continuing for as long as both source and target stay with range (the values of source and target revert to their original values once the function completes, however). All of this might sound very complex but it is a simple matter to specify, for instance, that a certain neuron should have some desired threshold and then call copy-to-range to set the same threshold to a whole bank of neurons. For example: .....9
(general neuron) 9999999999999
(neurocopy) 9........... This sequence specifies that neuron number 101 should have a threshold of 29 and then copies the same threshold to neurons 102 to 200. As it stands, it also copies axonal and dendritic connections, which might not be desirable, but if the sequence occurred early in the genome there might not be any connections at this point. (If it were not intended that connections be copied, a single digit 1, placed before the 3, 5 or 7 in the above Neurocopy gene, would toggle connection-copying off). Even if this gene mode is not used by a single human programmer, it is hoped that, like the Multiconnect mode, it will allow e-den evolution to create groups of neurons with similar functional capabilities that can work cohesively together, rather than having to evolve each neuron independently.
Major Gene Modes... Top... Contents... New Gene MarkerGene mode 9 is used to assign a value to the gene mode itself. Starting in Junk mode (gene mode 10), the first 9 sets the gene mode to 9 and thereafter the gene mode is set to value of the following digit. Thus, 9s themselves leave the syntax poised at the start of a new gene, and other digits introduce the specific gene modes discussed above. The logic determining transitions from genes to Junk and back was discussed above.Major Gene Modes... Top... Contents...
|