The segmented body pattern along the longitudinal axis of the Drosophila embryo is established by a cascade of genes coding specific transcription factors. This cascade is initiated by maternal gene products (BCD = bicoid and maternal HB = hunchback proteins) that are localized at the polar regions of the egg. The gap genes act at the top of this regulatory hierarchy. Expression of the gap genes occurs in discrete domains along the longitudinal axis of the embryo. Their protein products, which are DNA-binding transcription factors mostly of the zinc finger type, form broad and overlapping concentration gradients which are controlled by maternal factors and by mutual interactions between the gap genes themselves.
Once established, these overlapping gap protein gradients provide spatial cues which generate the repeated pattern of the subordinate pair-rule gene expression. The maternal gene products, in combination with various gap gene proteins, provide position-dependent sets of transcriptional activator/repressor systems which regulate the spatial pattern of specific expression of downstream genes, first of all pair-rule and segment polarity genes. Region-specific combinations of different transcription factors that derive from localized gap gene expression eventually generate the periodic pattern of pair-rule gene expression by their direct interaction with individual regulatory "stripe elements" of particular pair-rule gene promoters. Investigated in detail, the core gap genes containing part of the segmentation net are shown in figure 1.
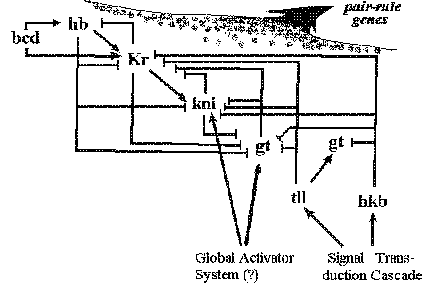
Figure1. bicoid (bcd) activates both hunchback (hb) and Kruppel (Kr) , hb activates and represses Kr in a concentration-dependent manner and represses the anterior limits of both knirps (kni) and giant (gt) expression, while the terminal gap genes tailless (tll) and huckebein (hkb) act as repressors of Kr , kni and gt . Tll is also required to activate posterior hb gene expression. Kni and possibly gt is thought to be activated by a global activator system for which no genetic basis is established (Jäckle et al. , 1992). Hkb and tll in turn are activated by the terminal signal transduction pathway.
It is well established that each segmentation gene has vast regulatory sequences containing multiple target sites for regulation of their activity. Multiple interactions of negatively acting transcription factors are directly responsible for setting the spatial limits of expression. But what is more, they probably do so by interfering with the cooperative binding of positively acting factors which react at nearby target sites. For example, the anterior limit of knirps gene expression is set by five or six interactions with regulatory sites to which the negatively acting HB protein binds, and the posterior limit requires multiple negative interactions at a different regulatory locus with the product of the gap gene tll (Pankratz et al., 1992).
My approach exploits the following essential characters of gene networks. Each gene-member of the network encodes a peptide-product whose function is activation or repression of another gene (its so called target gene). These transcription factors recognise specific sequences of DNA in regulatory domains of the target genes and tightly bind to the target sites. A given member of the network produces transcription factors that specifically activate or repress other netters, and vice versa. A complex network of autoregulatory and cross-regulatory actions functionally connects the genes thereby forming gene networks and cascades. The result of action of the network or cascade is a particular spatial pattern of activity (expression) of the network members. Down-activated members, in turn, switch on structural genes at the appropriate time and place. These genes, in turn, produce enzymes for the subsequent differentiation and morphogenesis of embryo rudiments.