A.V. Spirov
The Sechenov Institute of Evolutionary Physiology and Biochemistry,
Russian Academy of Sciences, St. Petersburg, Russia
INTRODUCTION
Homeobox-containing genes (homeobox genes) were first identified in the fruit fly Drosophila melanogaster. Protein products of these genes (homeoproteins) play a key role in control of embryogenesis [1-2]. Homeoproteins are site-specific transcription factors which regulate expression of target genes, particularly genes of other transcription factors (cross-regulatory action), and of intrinsic genes (auto-regulatory action). Auto-regulatory and cross-regulatory functional interactions integrate homeobox genes into gene networks. Apart from cross- and auto-regulatory interactions, expression of vertebrate homeobox genes is also regulated by retinoic acid. This acid functions through intranuclear retinoid receptors, such as RAR and RXR [5-6].
Each of the homeobox genes contains a highly conservative sequence of 183 pairs of bases (183 bp), the so called homeobox, which codes homeodomain, a sequence of 61 aminoacids. The homeodomain is responsible for recognition of homeoproteins and their binding to DNA. At present, several hundred homeobox genes are identified in various invertebrates and vertebrates. Peculiarities of the primary sequences of the homeodomains of the proteins-products make it possible to subdivide this large gene family into subfamilies (classes). The class of Antennapedia -like genes (the Antp-class) includes the homeodomains which at least by 60% are identical to the homeodomain that was first identified in a product of the Drosophila gene Antennapedia.
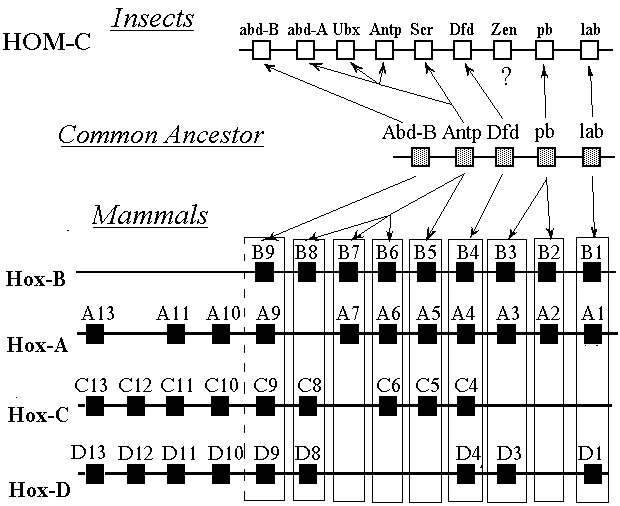
Fig. 1. Organization of the Hox-gene network in the human genome compared to organization of the HOM-C complex of the fruit fly Drosophila melanogaster (According to R.Krumlauf [2]).
Four gene complexes (clusters), HOX-A, HOX-B, HOX-C, and HOX-D are located relative to each other so that homologous genes of each family are in the same row. Vertical columns composed of homologous genes form subfamilies of Hox-genes. The total of 13 groups of paralogous genes are known. The Hox-gene names according to the modern nomenclature (see [9]) are written above the gene marks, while their names according to the old classification are indicated below the gene marks.
An essential peculiarity of the Antp-class consists in that its genes are grouped in chromosome clusters. Thus, all Antp-class genes in Drosophila are grouped in the so called homeotic complex (HOM-C complex) of the chromosome 3 [3]. Among vertebrates, the best studied are the murine ( Mus musculus ) and human Antp-genes grouped in four clusters, each cluster being located on its own pair of homologous chromosomes (Fig. 1). In the human genome, according to the recent classification, the Hox-families are located in the chromosomes 2, 7, 12, and 17 and are called Hox-A, Hox-B, Hox-C, and Hox-D, respectively [4]. In the mouse genome the Hox-families are located in the chromosomes 6, 11, 15, and 2 and are called in the same way as the human families (Fig. 1). The data obtained on Antp-genes in representatives of other vertebrate groups make it possible to claim with certainty that such organisation of the Antp-class structure (four independent clusters) is characteristic of the vertebrates in general.
Functions of Homeobox Genes
At present, an extensive information has been accumulated on the network of homeobox genes, which controls processes of determination of the vertebrate embryo axis organisation. The homeobox genes have been shown to be expressed in many embryonic patterns and structures [1, 2]. Comprehensive experimental studies on the gene expression pattern of all four vertebrate clusters allowed the conclusion that expression domains of each homeobox gene are clearly restricted [5], and such restriction of expression zones along the embryo axis is of prospective importance for the further embryogenesis. A definite correlation is revealed between the sequence in expression regions along the front-back axis and the physical cluster organization of these genes in the chromosome [6].
The term colinearity is accepted for this property, by analogy with a similar regularity that was first revealed by Lewis for Drosophila [7]. Such colinearity and a highly regular nature of the domain expression was observed in the mouse embryo for the nerve tube, neural cross, bronchial arches, extremities, mesoderm, and gonads (see [2]). The colinearity is a kind of code (Hox-code) for determination of the embryo regional specificity [8].
Evolution of the Antp-Class of Homeobox Genes
Homeobox gene networks could be particularly useful for studies on phylogenetic relations of evolutionary distant taxons, because the homeobox genes are characterised by conservatism within large evolutionary periods [10]. Besides, the presence of four copies of the ancestor cluster in each vertebrate species permits multiple comparisons of taxons, thus increasing the confidence level of phylogenetic analysis.
Ruddle's laboratory was one of the first to use similarity of the primary sequence of Antp-class homeobox genes to reveal phylogenetic relations within this group [10-12]. As a result, it was shown that there were two stages in the evolution of the vertebrate Antp-class homeobox genes. The first stage includes a gradual increase in the number of these genes within the ancestor cluster by a consecutive duplication of individual genes. (It is known that the more evolutionary ancient animal taxon is studied, the lesser number of the Antp-class homeobox genes is revealed in the cluster). The second stage is associated with duplications of the entire clusters (coupling groups). The process of evolution results in formation of a multigene family that is distributed among four clusters in four chromosomes. At each stage, the homeobox genes seemed to be under the press of selection, since the first stage is characterised by a quick divergence of initial homeobox sequences, whereas the second stage is much more conservative [10].
Organisation of Regulatory Sites of Homeobox Genes
In eucaryotes, according to the current viewpoints, the transcribed DNA molecule sites (the coding sequences, exons, separated by noncoding sites, introns) are separated by large intergene regions in chromosomes. Sites of the DNA molecule adjacent to the coding region from 5'- and 3'-ends (5'- and 3'-flank sites) as well as introns contain numerous specific sequences. Many of them are identified as regulatory elements. Each regulatory element, in turn, is a group (cluster) of closely localised target sites (or binding sites). The binding site is a small, highly conservative sequence of bases, which contains from 5 to 20 pairs of bases (5-20 bp) and is a target for recognition and specific binding by a transcription-regulating protein. The transcription factors are divided into activators and repressors. Target sites both for activators and for repressors are, as a rule, grouped within a cluster. Mechanism of action of the transcription factor that is binding with a target site remote from the promoter remains unclear [13].
Binding sites themselves are highly conservative, and so is the cluster organisation as a whole. However, the regions between the binding sites are highly variable, and their function is not clear. The regulatory sites of evolutionary distant but functionally similar genes may be characterised by little similarity of sequences as a whole and yet they may have target sites whose structure is identical (or rather similar) [14].
It is known that gene-regulating elements can be located in intergene regions rather far from their gene. Such organization of regulatory regions of eucariotic genes handicaps very much their search and analysis of phylogenetic relations. Homeobox genes are not an exception, and their regulatory elements are scattered at a distance of several thousand pairs of bases from the translation initiation site as well as in introns. However, a number of conservative elements in several large groups of the homeobox genes are located close to the starting site of translation, in the promoter region. These promoter regions of the homeobox genes have been deciphered for nearly 70 representatives of the Antennapedia class. Such extensive material undoubtedly is of much interest both for the analysis of the structure of the promoter regions and for the phylogenetic analysis of the homeobox genes.
Recent studies by Ruddle's group [10-12] have been dealing with the problem of phylogenetic relations in the family of Antp-like homeobox genes, based on the data on the homeodomain aminoacid sequence. As a result, the necessity is claimed of expanding research in this field and of analysing phylogenetic relations on the basis of regulatory sites of these genes. In the late 1980s, an obstacle for such studies was an insufficient amount of the genetic database. In the middle 1990s, a sufficient material for such analysis was accumulated, including that collected in our database, in particular (WWW http://www.iephb.ru/~spirov/hox-pro/hox-pro00.html). At present there is everything necessary to study this interesting phylogenetic problem.
CONSERVATIVE REGIONS IN HOX PROMOTERS