A.V. Spirov
The Sechenov Institute of Evolutionary Physiology and Biochemistry,
Russian Academy of Sciences, St. Petersburg, Russia
CONSERVATIVE REGIONS IN HOX PROMOTERS
In the homeobox genes, apart from homeoboxes, there are other conservative sites, including non-coding (structural and regulatory) regions (promoter region, introns, 3'-flank regions). This is specificity of modern methods of isolation of individual genes that in addition to transmitted sequences, the 5'-flank site is also deciphered. Its length does not usually exceed 300 bp, if to begin counting with the translation starting site. However, it is known that this 5'-flank site is functionally important and sufficiently conservative. Taking into account that the number of these decoded 300-800 bp promoter zones is comparable with the number of sequestered homeoboxes, the phylogenetic analysis of these zones definitely is as promising as that of 183-bp homeoboxes.
Using program packages such as CLUSTAL [15] and PHYLIP [16], we performed analysis of the promoter regions of the vertebrate genes of the Antp-class (Fig. 2). The promoter phylogenetic tree thus obtained resembles that for homeoboxes [11]. Analysis of dendrograms allowed several gene groups to be revealed on the basis of conservatism of sequences in the promoter zones. Two compact groups are clearly seen: genes-homologs of the Deformed Drosophila gene (Dfd) and homologs of the Sex comb reduced gene (Scr). The third group consists of homologs of Abd-A/Ubx/Antp Drosophila genes (Ubx-group; Fig. 2). The most evident and informative are examples of the homologous sequences in 5'-flank (promoter) regions of the homologous Scr- and Dfd-groups.
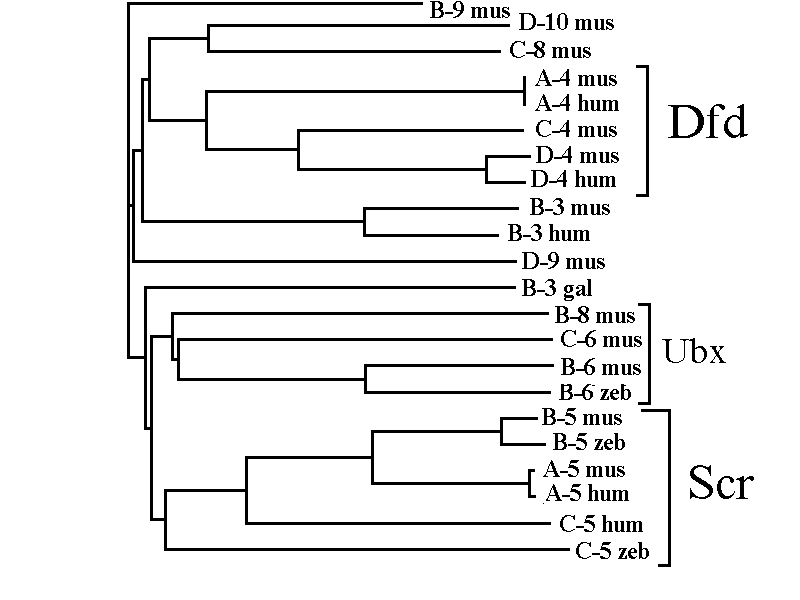
Fig. 2. The dendrogram of the promoter regions of the vertebrate Antennapedia -like homeobox genes.
The dendrogram is obtained using the program CLASTAL package [15]. The programs calculate indexes of similarity between all pairs of sequences which are later used for constructing the dendrogram. The method of mean relations is used for clusterisation of sequences. Clusterisation of the Dfd-, Scr-, and Ubx-groups is noticed. After the gene name, the source is indicated: hum, human; mus, mouse Mus musculus; gal, hen Galus galus; zeb, zebrafish Danio rerio.
To consider this problem in detail, the comparative phylogenetic analysis of homeodomain sequences and sequences of the promoter zones will be done only for the Dfd- and Scr-groups. Genes of these groups are quite likely to be characterised by a self-activation by its own homeodomain-containing protein-product [17-19]. Therefore, of special interest is comparison of dendrograms of promoters and homeodomains for these genes. Results of the phylogenetic analysis of the homeodomains of proteins-products of the Dfd- and Scr-group genes compared with the sequences of the first 300 bp above the triplet of the translation start are presented in Fig. 3. The dendrograms for the homeodomains and for promoter regions prove to be quite similar. On both dendrograms, the Dfd-group forms one arm, while the Scr-group, the other.
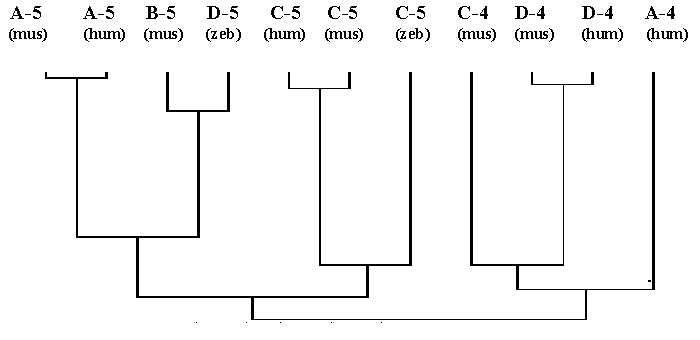
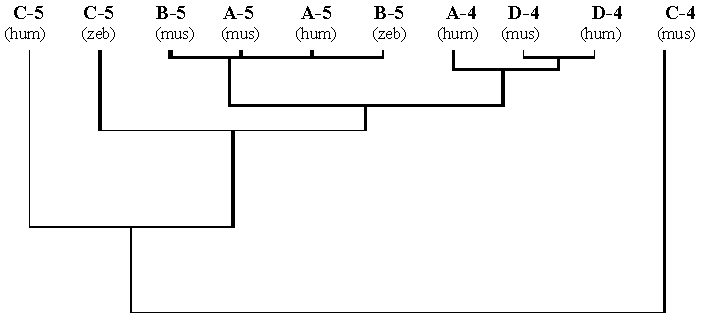
Fig. 3. The dendrogram of the promoter regions of vertebrate Antennapedia -like homeobox genes from the Dfd- and Scr-groups (A) compared with the dendrogram of homeodomains of the proteins-products of the same genes (B).
The same methods are used as those for the dendrogram in the Fig.2. See details in the text.
The Promoter Zone of Scr-Group
The 5'-flank zone seems to be the most conservative in the Scr-group. Conservatism of the sequences in the promoter zone of this group is noted by a number of authors [17, 20-22]. Homology of 5'-flank regions as a result of conservatism in the structure of a local regulatory element is illustrated by the example of the Scr-group (Figs. 4, 5). This element controls sensitivity of expression of Scr-group genes to intranuclear receptors of retinoic acid and to homeoproteins. It is essential that the activity of these elements is also confirmed experimentally [17, 21].
box b box c
_______ ______________________________
HoxA-5(mus) .cccccattagt.GCAC.....GAGT.....ttacctCTAGaggtcaTCAGGCAGGATTTACGACTGG
HoxA-5(hum) .c-cct--t-gt.-C-C.....---T.....--------A--g--------GC--G-----------G
HoxB-5(mus) at-gtc--t-GGT-T-ACCATA---CATGAA--------T--g--------CG--A-----------.
HoxB-5(zeb) CT-GTC--C-TTT-T-ACCATA---CATGAA--------T--A--------TG--A-----------G
box d
____________________
HoxA-5(mus) ACAACAAAAGCACGTGATTC..
HoxA-5(hum) A-------------------..
HoxB-5(mus) T-------------------CC
HoxB-5(zeb) T-------------------TC
Fig. 4. Results of the search and analysis of conservative elements, with consideration of multiple alignments of the conservative site in the promoter regions of the mouse and zebrafish ( Danio rerio ) HoxB-5 gene (hox-2.1), and the mouse and human HoxA-5 gene (hox-1.3) (using the programs CLASTAL).
Point means the local break in the aligned sequence; dashes mean identity of the nucleotide in the column. The known and proposed sites for the action of transcription factors are marked by lower-case letters. ccccattagt (box b) and atcgtcatta are the known binding sites for the transcription factor HOX-1.3 and for the protein-products of the homeobox genes of Antp-class, respectively [17]. ttacctNNNNaggtca is the proposed target site for the action of intranuclear retinoid receptors (N - any nucleotide). Sites marked as box c and box d bind specifically unidentified transcription factors.
The presence of binding site of the protein-product of the HoxA-5 gene (hox-1.3)* in the promoter zone of this very HoxA-5 gene (a gene self-activation element; Fig. 4) is experimentally shown by Odenwald [17]: the same protein HOX-1.3 was found to have a binding site in the promoter zone of the HoxB-5 gene (hox-2.1; Fig. 4). When compaing base sequences in the human HoxC-5 gene (hox-3.4) and its homolog in the zebrafish Danio rerio, of the greatest interest is the promoter element 180 bp which turned out to be homologous by 60% in these two genes [22]. Arcioni et al. [21] have found that the promoter zone discussed contains one site for binding the retinoic acid receptor (box A) and two sites for binding homeoproteins (box B and box C; Fig. 5). Comparison of the human gene HoxC-5 promoter with homologous mouse genes HoxA-5/HoxB-5/HoxC-5 showed the regulatory elements of the mouse and human HoxC-5 to be actually identical, whereas more remote HoxB-5/HoxA-5 homologs contained less close sequences in all three sites [21]. On the one hand, comparison of the zebrafish HoxC-5 promoter with the human HoxC-5 promoter revealed that both binding sites of the homeoproteins (box B and box C) were conservative, which indicates a functional conservation of these regulatory elements. On the other hand, the retinoic acid binding site (box A) receptor is significantly less conservative, so that the 10 bp-palindrom sequence is reduced to the 6 bp site in the zebrafish Hox-5 gene.
box B ___________________ HoxC-5(hum) GGAGGTCATCAAGCCAAATTTATGAGTGGCC HoxC-5(mus) GGAG.TC----------A-------G----- HoxC-5(zeb) GGGG...----------.-------C----- box A _____________________ HoxC-5(hum) GCTCgagtcacgtgactcTATTTAAGGCTCCCT << >> HoxC-5(mus) GCTCCAGT-------C---------GGC-C--T << >> HoxC-5(zeb) AGGAGCTG-------T---------ACA-T--A << >> box C ________________ HoxC-5(hum) TA.........TCTCCACCTATAAATTG HoxC-5(mus) TA.........T--------------TG HoxC-5(zeb) GAAAATGATTTC--------------CC
Fig. 5. Results of analysis of conservative elements, taking into account multiple alignments of the conservative site in the promoter regions of the human, mouse, and zebrafish ( Danio rerio ) HoxC-5 gene (hox-3.4).
Three regulatory sites are marked: box A (target palindrom for the action of retinoid receptors) and targets for the action of the homeoproteins - box B and box C [21-22].
Promoter Zone of Dfd-Group
Nearly 300 first base pairs in the 5'-flank zone of vertebrate genes whose homeobox structure is homologous to that of the Deformed Drosophila gene demonstrate a high similarity between each other (Fig. 6). Unfortunately, it is on these sites of the promoter region that there are no experimental data in the literature with respect to their functioning sites of binding of transcription factors. However, there are such experimental data on two regulatory elements with a great distance from triplet of the translation start: the auto-regulatory element and the element for the action of retinoic acid receptors [23, 24-25].
HOX-1.3 ________ HoxD-4(mus) ctctattaGCATCTGTCAGGGACTCTCAAATGTGGCATGGCAAGTCACTTGATT HoxD-4(hum) ctacattaAT-T-TGG-AGGGG-T-T--AATGTGC--TAGCA--CTA-TTG-TT HoxC-4(mus) AGCTTCTGCC-G-CAT-CCCCA-C-C--CCCCCAG--GCAGC--CAC-ACC-CC RARs _______________ HoxD-4(mus) ACACGTATGTTAT.TTAGTTAAATTTGTGAAAATTATGAGATGCtcaccAACCCggtgaT HoxD-4(hum) ---CGT-TGTT-T.--A-T--A----G----A-TTAT--GAT-CtcaccA-C--ggtgaT HoxC-4(mus) ---TAC-CAAA-AA--G-A--C----.----T-AAGC--TTC-GTTCCTT-T--GGGGAC "ccaat" _____ HoxD-4(mus) AAACTTGCTTTCTTCCTattggCT HoxD-4(hum) AAACT----CCC-CGCC------T HoxC-4(mus) TGGGT----CGG-.GTG-----CC
Fig. 6. Comparison of sequences in the beginning of the promoter region of the human and mouse HoxD-4 (hox-4.2) gene and of the mouse HoxC-4 (hox-3.5) gene.
Proposed sites for the action of transcription factors are marked by lower-case letters: ctacatta - binding site of the transcription factor HOX-1.3; ccaccNNNNNggtga - target site for the action of intranuclear retinoid receptors; attgg - binding site for proteins of a large group of ccaat -factors of transcription .
--------------------------------------------
It should be noted that by using computers, in 5'-flank zone the sites were revealed which are potential targets for the action of the transcription HOX-1.3 factor in the promoters of human and mouse HoxD-4 (hox-4.2) genes (Fig. 6). And the homologous murine HoxC-4 gene (hox-3.5) contains in this place a potential binding site of another homeoprotein, BICOID. Moreover, an obvious similarity is observed when comparing the experimentally established structure of the promoter zone of the Scr-group genes with results of computer scanning of the Dfd-group genes (compare Figs. 4-5 and 6). In both cases, the binding site of the homeoprotein (particularly, the HOX-1.3 factor) is followed by the binding site of the retinoic acid receptor (3'-direction). It is essential that in the promoter zone of the human and mouse HoxD-4 gene there is a palindrom (Fig. 6), and its semisites, according to their sequence, are appropriate for binding retinoic acid receptors [26] and are similar to target-palindrom of the retinoid receptor in the rGH-promoter [27].
It is shown experimentally [24] that there is a functioning element for the action of retinoic acid receptors (particularly, the RAR factor). It is located far from the translation starting site of the mouse HoxD-4 gene, in its 5'-flank region. Meanwhile, not far from this regulatory element, an auto-regulatory element of the HoxD-4 gene is located [25], its activity being determined by two target sites for binding of homeobox proteins. The receptor element for retinoic acid, which is similar in localization, structure and functions, is found in the promoter zone of the human HoxD-4 gene [23]. It is also essential that gene-engineering experiments showed the presence of a similar regulatory element for retinoid receptors in the 5'-flank region of another Dfd-group representative, the mouse HoxA-4 gene (hox-1.4) [28]. Moreover, recently, the presence of auto-regulatory elements for the HoxA-4 gene has been shown [19].
On the other hand, it is noted in the paper [24] that the initial sequence of the promoter zone of the HoxD-4 gene, the first 395 bp, were not tested in gene constructs; meanwhile, it is in this region that the additional sites of the RAR action might be located, which can be substantiated by the data on activation kinetics. Therefore, it is reasonable to believe that the binding sites of retinoid receptors revealed by scanning in the beginning of the promoter region of Dfd-class genes (Fig. 6) might be these missing elements.
DISCUSSION: EVOLUTION OF HOMEODOMAINS AND REGULATORY ELEMENTS